Infrastructure-as-code in the life sciences
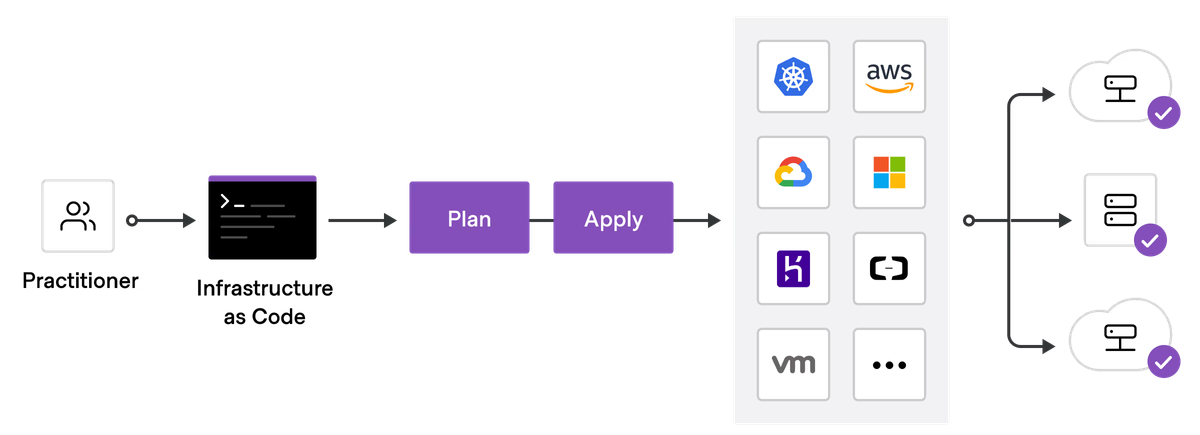
TL;DR - Don't use the cloud console to manage your cloud infrastructure. Instead, implement infrastructure-as-code (IaC) tools like Terraform or OpenTofu. Or use a Platform-as-a-Service designed for the life sciences like Ganymede. The life sciences are particularly prone to manually configured infrastructure because bioinformatics setups form a simple "base" of where many companies start their tech stack with file-storage buckets and basic compute VMs or batch compute tools, but this approach is poorly suited to scaling in later stages.
Getting started with the cloud is easy. You create an AWS or GCP account using your email, follow a tutorial that guides you through all of the needed UIs, then sit back and show off your work to all your colleagues. Look at all the data in that S3 bucket!
Then your co-workers request some permission changes, someone has to make a few updates while you're out on vacation, and now the Process Development team also wants the same setup for their bioreactors (with just a few "small" tweaks.) Do you remember all of the changes you made across months to get things set up? Do you have enough focus time to work with two open tabs and manually recreate a new setup based on clicking through each page of the working configuration in the console? How do you update your setup to include a streaming data store since bioprocess data isn't really file-based?
The cloud console is excellent for learning and prototyping, but it's not designed for simplicity, repeatability, or scalability. That’s why infrastructure-as-code (IaC) tools exist and can overcome the cloud console's biggest challenges.
Complexity rears its head
You shouldn't be using the cloud console to manage your infrastructure
Console complexity
Most cloud tutorials start by walking through steps in the console, which is perfect for introducing concepts to new users. Once you start building things beyond the "hello world" configurations however, the complexity rapidly piles up. Look at this tutorial for connecting an S3 bucket across AWS accounts: it has 19 pre-requisite steps and more than 40 steps that bounce between the web UI, CLI, and links to other tutorial pages. In addition to the time spent running through these steps for each bucket, the time spent learning and understanding the same steps for every other configuration change poses a significant drag on execution. You'd be better off using this time interviewing your scientists and bioinformaticians to learn their needs and building business/pipeline logic and UIs for them.
Repeatability
Most software efforts start with a single environment, which is a single copy of all of the components needed to implement a solution. Let's imagine we're setting up a private python package repository and S3 bucket on AWS that can be reached from a virtual machine to do some basic analysis. We'll need to create a AWS project, enable the necessary services, create the resources, configure authentication, etc. Bioinformatics pipelines are often some of the first infra that biotechs will create, and look like this. Once that's been debugged and is up and running, things will run smoothly – until a change to the python package introduces a bug in a workflow.
Once that's resolved, a likely outcome is to add some pre-release testing to the python package using a dev or staging environment. To set this up you'll need to recreate each piece of infrastructure in a new project, and ensure that it matches the configuration of the production environment going forward. Repeating these setup steps are time consuming not only in initial execution (see console complexity), but also in troubleshooting and ongoing maintenance.
As your pipelines and apps scale from exploratory one-offs used by few people to larger organization-wide tools and full software applications, and your science also coalesces into more of a PD/AD phase, reproducibility is key for scaling, quick recovery, and ultimately also helping to lock down your app as you approach more regulated phases of development. The infrastructure also mutates quickly over time as you move from batch file-based compute suited for bioinformatics or machine learning pipelines, into more advanced transactional applications or robust analytical workflows. IaC can help you keep up with these changes.
Wasted effort
Biotech companies aren't formed to become experts in cloud configuration and many early-stage companies have limited headcount to invest in software development and infrastructure engineering. Hiring great minds in bioinformatics and the sharpest automation engineers, and then asking them to read through AWS Role JSON files for misconfigurations, is neither beneficial to the enterprise nor the employee.
As more time is spent on debugging and toilsome cloud console work, the team's energy will drain and a palpable feeling of misdirected energy will arise. Biotechs have a clear mission to improve the world through the cutting edge of science – and the more time that is spent focused on that mission, the higher the likelihood of success.
Moving away from the console
Setting up IaC isn't a painless process, as it involves some initial cloud console configuration (you can never escape it totally 😀), setting up code repositories, and some deployment process design. Ideally every change to IaC is linted, tested, and code-reviewed prior to deployment (it's code after all) and then run through a continuous deployment (CD) pipeline so that all of your development, staging, and production environments are up to date.
Or you could skip the challenges of the cloud console and vault to the next level of maturity by using an internal developer platform like Ganymede that not only abstracts away the infrastructure, but also the IaC, and offers a fully prebuilt environment for connecting your lab's data, apps, and automating analysis. Interested in learning more or just picking our brain about infra-as-code for your biotech? Reach out at hello@ganymede.bio!

